Biology

EMODnet Biology provides open and free access to interoperable data and data products on temporal and spatial distribution of marine species (angiosperms, benthos, birds, fish, macroalgae, mammals, phytoplankton, reptiles, zooplankton) and species traits from European regional seas, as defined by the EEA’s 'Europe’s seas’ dataset (Arctic Ocean, (North) Atlantic Ocean, Baltic Sea, Black Sea, Mediterranean Sea and North Sea).
EMODnet Biology's taxonomic backbone is built upon the World Register of Marine Species (WoRMS ) and supported by the European Ocean Biodiversity Information System (EurOBIS ) data infrastructure, with tools and services developed in collaboration with Lifewatch ERIC and Lifewatch Marine.
Because EMODnet Biology is OGC compliant, it enables access to metadata descriptions of more than 1,200 thematic biological datasets. Due to being INSPIRE compliant these metadata records can also be found through the EU Open Data Portal. Our data and metadata follow the FAIR principles and we continuously work on improving the Findability, Accessibility, Interoperability and Reusability for all records and data.
Objectives
EMODnet Biology aims to provide a single access point to European marine biodiversity data and products collected in the European regional seas, as defined by the EEA’s Europe’s seas dataset as well as the Caribbean Sea. Through our interoperable products, created by assembling individual datasets from various sources, we contribute to the environmental state of ecosystems and sea basins’ assessments.
The main objective is to contribute to EMODnet’s operational service by maintaining and enhancing services for the European biodiversity data and products.
EMODnet Biology’s specific objectives are defined in the following tasks:
- Task 1: Maintain and improve a common method of access to data held in repositories;
- Task 2: Construct products from one or more data sources that provide users with information about the distribution and quality of parameters in time and space;
- Task 3: Develop procedures for machine-to-machine connections to data and data Products;
- Task 4: Contribute data, data products and content to EMODnet Central Portal that allows users to find, view and download data and data products;
- Task 5: Contributing static content to dedicated spaces in the Central Portal;
- Task 6: Ensure the involvement of Regional Sea Conventions (RSC)
- Task 7: Contribute to the implementation of the EU legislation and broader initiatives for open data;
- Task 8: Monitor quality/performance and deal with user feedback.
Background
Europe’s seas and oceans are home to a staggering abundance and diversity of life, from large charismatic species such as seals, whales and dolphins, to the microscopic marine algae that form the base of the marine food chain. More than 36,000 known species of marine plants and animals are found in Europe, and understanding their geographic distribution, abundance and seasonal, annual or decadal variation is key to detecting changes in the marine ecosystem and for assessing ecosystem health of maritime basins. Unfortunately, measuring or observing marine life on a large scale is difficult.
Marine biodiversity data are essential to measure and study the ecosystem health of maritime basins. These data are often collected with limited spatial and temporal scope and are scattered over different organisations in small datasets for a specific species group or habitat. In addition, as data are collected by multiple organisations, using different standards, technologies and conventions, it is challenging to combine them. Furthermore, a plethora of historical marine biodiversity datasets exist in the form of simple and unorganised printed documents or electronic files, on the hard disks or other media of electronic information storage of individual scientists and of marine institutes, research centres, academic departments, ministries, port authorities, public or private libraries. These types of data, which are not stored on a remote server (e.g. in the cloud or a research repository), are considered to be at permanent risk of being lost to future use. It is these datasets, however, which provide the historical context for present observations, facilitating the establishment of reference conditions for monitoring and management.
History of EMODnet Biology
The Maritime Policy Blue Book, welcomed by the European Council in 2007, announced that the European Commission would take steps to set up a European Marine Observation and Data Network to improve access to high quality marine data for private bodies, public authorities and researchers.
The consortium comprises 23 organisations, with four partners qualified as IODE Associated Data Units (ADU) and ten others as IODE National Oceanographic Data Centres (NODC). The partnership is geographically distributed and includes all European OBIS nodes (EurOBIS, MedOBIS , OBIS UK , OBIS Black Sea ). Collaboration with other international initiatives is also ensured via the consortium with the inclusion of e.g., IODE and ICES.
This phase will see an expansion of our web services, the implementation of management practices for other types of data like -omics and images and the migration to the Central Portal. Several new data products will be created and made freely available and a closer engagement with our stakeholders will ensure that our work will better answer their requirements.
The consortium is composed of 23 organisations with nine partners qualified as IODE National Oceanographic Data Centres (NODC) and four as IODE Associated Data Units (ADU). As in previous Phases, the partnership is geographically distributed across Europe and includes all European OBIS nodes (EurOBIS, (EurOBIS, MedOBIS , OBIS UK , OBIS Black Sea ). Collaboration with other international initiatives is also ensured via the consortium with the inclusion of e.g., IODE and ICES. Collaboration with other OBIS nodes, technical, data republication via EMODnet Biology is ensured via informal contacts. The collaboration currently focuses but is not limited to AntOBIS (Antarctic node), Caribbean OBIS , OBIS Deep Sea , OBIS SeaMap , OBIS Kenya and Oceans Past Initiative .
Following the centralisation work that was completed in January 2023, this new Phase will focus on maintaining and improving our webservices so users can access our data and products. We will also continue to develop our data infrastructure to accommodate for the publication of new types of data and will have a number of actions to better engage with our stakeholders as well as create products that can best address their needs.
Partnership
The reach and breadth of the EMODnet Biology consortium represents a high-level of connectivity at the national, regional and global scales. An overview and details of the Phase IV partnership can be found in the Partnership page.
As part of an exercise, in Phase IV (2021-2023), to better understand the linkages we have with key stakeholders and initiatives, we have created a map with the connections and touchpoints. This was done, following a targeted questionnaire to EMODnet Biology partners, we then used the HighCharts application (see below) to provide a visualisation of the key linkages with UN Decade Programmes, Regional Sea Conventions, ICES Working Groups and the other EMODnet thematic lots. In Phase V we plan to update this connectivity map with the current consortium and add other relevant connections to initiatives and/or programmes.
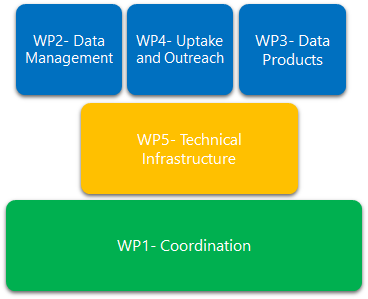
Work Packages (WP)
Phase V started in May 2023 and will run until May 2025, the workplan is divided in five work packages, as follows:
- WP1: Coordination (lead partner: VLIZ);
- WP2: Data Management (lead partner: VLIZ);
- WP3: Data Products (lead partner: INRAE);
- WP4: Uptake and Outreach (lead partner: OGS);
- WP5: Technical Infrastructure (lead partner: VLIZ).
Objectives
WP1 is set up to provide support to the whole project in terms of coordination and ensuring that all deliverables and objectives are met within the timelines proposed in the tender and also in this proposal. This WP will focus on Task 8: Monitor quality/performance and deal with user and is also responsible for organising project meetings with all parties involved, attending the EMODnet network meetings, as well as other events, e.g. conferences, workshops, seminars, etc., for which an invitation is received or participation required. Another main objective is ensuring that reporting is submitted according to the tender specifications, with guidance provided by the EMODnet Secretariat.
Methodology & activities
In order to achieve the objectives as included above, several Actions are planned for Phase V:
- Action 1.1 Usage of service monitoring via indicators (Lead: VLIZ);
- Action 1.2 Maintenance of centralised EMODnet catalogue of products (Lead: VLIZ, Partner: INRAE);
- Action 1.3 User feedback and support to the Centralised EMODnet helpdesk (Lead: VLIZ);
- Action 1.4 EMODnet Biology consortium meetings (Lead: VLIZ, Partners: INRAE, OGS);
- Action 1.5 EMODnet meetings/events (Lead: VLIZ, Partners: INRAE, OGS);
- Action 1.6 End of Phase Handover (Lead: VLIZ).
Deliverables
- D1.1.1 Quarterly Progress reports (M4-M24);
- D1.1.2 Interim report 10 May 2024 (M12);
- D1.1.3 Final Report 10 May 2025 (M24);
- D1.2.1 Update of EMODnet Biology data products via the EMODnet CP catalogue (M12, M24);
- D1.3.1 Monitoring user feedback via EMODnet JIRA (M0-M24);
- D1.3.2 Operation of the EMODnet CP helpdesk (M0-M24);
- D1.4.1 Minutes from the three project meetings (kick off, annual and final) (M1, M12, M24);
- D1.5.1 Participation in the EMODnet Steering Committee meetings (M0-M24);
- D1.5.2 Participation in the EMODnet Open Conference (M0-M24);
- D1.6.1 Appropriate mechanisms and guidelines for handover (M24).
Objectives
The main objective for WP2 is defined by Task 1- Maintain and improve a common method of access to data held in repositories, which will be addressed through the planning of various actions.
Methodology & activities
All efforts, data and outcomes of all previous phases of EMODnet Biology will feed into Phase V and the outputs planned will be split in five main Actions:
- Action 2.1 Data interoperability (Lead: VLIZ; Partners: OGS; Subcontractors: Aarhus, Deltares, HCMR, ICES, IEO, IH Cantabria, ILVO, IMR, IPMA, MARIS, MBA, NIOZ, Pangaea, SMHI, SYKE);
- Action 2.2 Streamline data holdings through Cloud technologies (Lead: VLIZ)
- Action 2.3 Evaluation of data restrictions (Lead: VLIZ; Partners: OGS; Subcontractors: Aarhus, Deltares, HCMR, ICES, IEO, IH Cantabria, ILVO, IMR, IPMA, MARIS, MBA, NIMRD, NIOZ, Pangea, SMHI, SYKE)
- Action 2.4 Data training workshops (Lead: HCMR; Partners: VLIZ)
- Action 2.5 Publication of omics data (Lead: VLIZ)
Output (Deliverables)
- D2.1.1 Maintenance of adequate mechanisms to ensure data are interoperable (M0-M24);
- D2.1.2 Report on the standardisation and integration of datasets published during the Phase (M24);
- D2.2.1 Summary on consortium data flows (M6);
- D2.3.1 Plan to optimise procedures to make restricted data available to users (M0-M24);
- D2.4.1 Workshop on use of Zooniverse for data digitisation through citizen science (M6);
- D2.4.2 Data training workshop for Mediterranean organisations (M6);
- D2.4.3 Report on the availability of data following the workshops (M22);
- D2.5.1 Guidance for data management practices applied to omics data (M0-M24).
Objectives
WP3 is set up to address Task 2: Construct products from one or more data sources that provide users with information about the distribution and quality of parameters in time and space. Building on previous Phases, WP3 will continue to create distribution maps targeting groups of species, as well as presence/absence maps, both of which can be queried, subset and downloaded via the EMODnet Central Portal viewer.
WP3 will develop efforts to maximize the uptake of existing and new data products by end users. This will be achieved by raising awareness of existing products, provide training materials in form of tutorials to manipulate and visualize EMODnet products using the existing R packages developed in Phase III and IV, and by co-creation of data product needs with stakeholders. The tender also requires the creation of products targeting species migration routes, in particular seabirds and cetaceans. Although there have been specific efforts made to increase the amount of data in EMODnet Biology for these two groups of species, the volume of data is still considered too limited to create sound and robust products for these taxa. Another constraint is that the current state-of-the-art for modelling distributions of these taxa uses individual movement data and highly sophisticated statistical analysis that is beyond the scope of this tender. Specific new actions will be taken to increase the data availability of these taxa by involving experts, by assessing the data sources of existing external products and indicators, and assessing the possibility of hosting these external products in EMODnet Biology.
During Phase V, a number of changes were implemented concerning data and product formatting, owing to the centralisation of the EMODnet services. Guidance for this was provided to all WP3 members on how to create their products in order to facilitate their incorporation in the EMODnet Central Portal Viewer. In Phase V we will continue with this approach and will adjust the guidance following the input of the EMODnet Secretariat, Central Portal team and any decisions resulting from the EMODnet Steering Committee.
Methodology & activities
In order to achieve the expected outputs, the work will be split in three main Actions:
- Action 3.1 Evaluation of data products from previous Phases (Lead: INRAE; Partner: OGS, VLIZ; Subcontractors: Cefas, Deltares, ICES, IH Cantabria, ILVO, MBA, NIMRD, NIOZ, SMHI, SYKE, UkrSCES, ULiège, U Sheffield);
- Action 3.2 Community calls (Lead: INRAE; Partner: VLIZ; Subcontractors: Cefas, Deltares, IH, Cantabria, NIOZ, SMHI, ULiège, U Sheffield);
- Action 3.3 Data product creation (Lead: INRAE; Partner: VLIZ; Subcontractors: Cefas, Deltares, IH Cantabria, NIOZ, SMHI, ULiège, U Sheffield).
Deliverables
- D3.1.1 Internal WP3 and WP4 Workshop 10 Sep 2023 (M4);
- D3.2.1 Quarterly community calls (M3-M24);
- D3.3.1 Liaison with external entities targeting seabird and cetacean migration outputs (M0-M24);
- D3.3.2 Data product creation (M0-M24).
Objectives
WP4 will cover three Tasks, more specifically: Task 5. Contributing static content to dedicated spaces in the Central Portal, Task 6: Ensure the involvement of Regional Sea Conventions RSC and Task 7: Contribute to the implementation of the EU legislation and broader initiatives for open data. Building on the expertise and connections developed in previous Phases, several actions will be put in place to ensure that the Tasks are achieved.
Methodology & activities
The Actions proposed are as follow:
- Action 4.1 Provide and update content to the EMODnet Central Portal (Lead: VLIZ; Partner: OGS; Subcontractors: ICES, ILVO, MBA, NIMRD, SYKE);
- Action 4.2 Interaction with the RSCs and ICES (Lead: OGS; Subcontractors: ICES, ILVO, NIMRD, SYKE);
- Action 4.3 Liaison with relevant EU initiatives, projects and parties (Lead: OGS; Partner: VLIZ; Subcontractors: ICES, ILVO, MBA, NIMRD, SYKE);
- Action 4.4 Ensure compatibility with INSPIRE Directive (Lead: OGS, Partner: VLIZ).
In previous Phases various lines of work were established to ensure that EMODnet Biology assets, e.g. metadata, data and data products are compliant with the INSPIRE directive. In Phase V the proposed work will be towards a better engagement with EU Member States, more specifically in providing support with their reporting obligations regarding, for example MSFD indicators.
Deliverables
- D4.1.1 Update connectivity maps and identification of stakeholder community (M4) ;
- D4.1.2 Informative material based 4.2.2 published in the EMODnet CP (M12);
- D4.1.3 Informative material based on the 4.2.3 outcomes (M14);
- D4.1.4 Informative material based on the 4.3.1 outcomes (M24);
- D4.1.5 Publication of four Use Cases (M6, M12, M18, M24);
- D4.1.6 Publication of written documents (M0-M24);
- D4.2.1 Engagement plan for each RSC (M6);
- D4.2.2 Questionnaire to inform about what data & products EMODnet Biology offers and to collect stakeholder needs (M6);
- D4.2.3 Workshop with RSCs to understand major needs (M12);
- D4.2.4 Report on progress for the publication of fisheries survey data (M24);
- D4.3.1 Workshop with representatives of major recent and current European Research Projects and initiatives to share experience and knowledge in biodiversity monitoring and assessment and in data, tools and services (M18);
- D4.3.2 Participation in TG DATA meetings (M0-M24);
- D4.4.1 Ensure compatibility with INSPIRE Directive (data, metadata, data products) (M0-M24);
- D4.4.2 Report on the activities to support EU Member States regarding their reporting obligations (M24).
Objectives
WP5 is responsible for fulfilling Task 3: Develop procedures for machine-to-machine connections to data and data Products and Task 4: Contribute data, data products and content to EMODnet Central Portal that allows users to find, view and download data and data products, as required in the tender document. VLIZ is the WP lead and will be supported with the technical capacity of other organisations (some already identified and others will be identified once the contract is signed) for accomplishing the objectives therein. The work expected for Task 3 should result in the maintenance and further development of Application Programme Interfaces (APIs), such as web services, to allow for metadata, data and data products to be found, viewed and downloaded by applications running on other machines. An alignment of the plan and solutions with the other thematic lots should be achieved to abolish barriers for users and operating systems. The plan defined for Task 3 shall be discussed during the EMODnet Steering Committee meetings and validated by the Contracting authority before it can be implemented. Within Task 4, the main objectives focuses on ensuring that metadata are easily discoverable for cataloguing and INSPIRE services, that data and data products can be discovered, viewed, downloaded and accessed by EMODnet Central Portal in a straightforward manner and with minimum steps and to ensure that coherence with the other thematic groups is achieved by participation in regular meetings of a Technical Working Group.
Description of work
Four Actions are planned for this WP and their detailed description is as follows:
- Action 5.1: Extend interoperability and data types (Lead: VLIZ, Subcontractor: MARIS);
- Action 5.2: Implement machine-to-machine connections to data and data products (Lead: VLIZ);
- Action 5.3: EMODnet Biology Technical Infrastructure maintenance and operational management (Lead: VLIZ);
- Action 5.4: Contribute data, data products and content to EMODnet Central Portal that allows users to find, view and download data and data products (Lead: VLIZ).
Deliverables
- D5.1.1 Streamlining semantic interoperability (M0-M20);
- D5.1.2 Integration of omics data into the data model (M16);
- D5.1.3 Roadmap for marine biodiversity data (M24);
- D5.2.1 M2M communication plan and solutions (M0-M24);
- D5.3.1 Operational web services (M0-M24);
- D5.3.2 Technology stack upgrades (M24);
- D5.3.3 Technical maintenance of procedures for ingestion of data from the data providers (M0-M24);
- D5.3.4 Development and implementation of procedures to publish omics data (M24);
- D5.4.1 Synchronisation of all data and data products metadata to the EMODnet CP catalogue (GeoNetwork) (M0-M24);
- D5.4.2 All data and products available through the EMODnet CP (M2-M24);
- D5.4.3 Participation in the EMODnet TWG meetings (M0-M24);
- D5.4.4 Maintenance and development of API used to query EMODnet Biology layers available via the EMODnet CP viewer (M0-M24).
Key services
EMODnet Biology provides key services and products which allow users to search and visualise data and related data products:
- Occurrence data (species observations) as Web Feature Service (WFS); Additional measurements, linked to the occurrence data as Web Feature Service (WFS);
- Retrieve data of the gridded abundance data products as WFS/WMS;
- Using the AphiaID to query by (biological) taxonomy;
- Using the IMISDasID to query by metadata dataset;
- Examples of EMODnet Biology Data applications written in R.
The Integrated Publishing Toolkit (IPT) , a freely available open source web application using the Darwin Core standard, will make it easy to share biodiversity-related data and information with the EMODnet portal.
The Quality Check (QC) tool was created to support data providers in assessing the quality of their datasets prior to submission. It can be used either through an Rshiny application or as an R package. Please check our video tutorial on how to use this tool and understand its output.
Training originally set up for EMODnet Biology Partners, opened up for self-enrolment since the November 2023. It allows access to scientists & data managers to use the available material to learn how to format, standardise and quality control their biodiversity data for submission to EMODnet Biology.
You can enrol and follow the course on OTGA (Ocean Teacher Global Academy): classroom.oceanteacher.org/enrol/index.php?id=958
The R package , partially funded by EMODnet Biology, is designed to make all EMODnet thematic lots’ raster data layers easily accessible in R. The package allows users to query information on and download data from all available EMODnet Web Coverage Service (WCS) endpoints directly into their R working environment.
The R package , created by EMODnet Biology partners, is designed to make all EMODnet thematic lots’ vector data layers easily accessible in R. The package allows users to query information on and download data from all available EMODnet Web Feature Service (WFS) endpoints directly into their R working environment.
BTrait was developed in the scope of EMODnet Biology to facilitates working with species density data, combined with species traits in R. It allows users to query the linked datasets (species density and trait data) and visualise it with an interactive shiny application.
The following file includes information about the mandatory and recommended fields for data submission to EMODnet Biology. Please contact us via bio@emodnet.eu if you have any questions.
Data and Products
EMODnet Biology provides access to data from a wide range of sources and actively pursues the inclusion of new and historical datasets to the inventory based on careful assessment of the ease of use and fitness for purpose of the data and associated databases.
The databases feeding into EMODnet Biology contain data from all regional and sub-regional seas of Europe, as specified by the Marine Strategy Framework Directive.
To ensure interoperability, EMODnet Biology implements (and if necessary adapts) common standards and vocabularies defined and used by SeaDataNet, WoRMS (World Register of Marine Species), OBIS (Ocean Biodiversity Information System), INSPIRE, GBIF (Global Biodiversity Information Facility), Marine Regions and the Lifewatch infrastructure.
Data sources
The main data contributors are:
- International biogeographic datasets from EurOBIS (European Ocean Biogeographic data system);
- National monitoring programmes;
- International monitoring campaigns (databases storing data from multiples countries within the same regional European sea);
- International data aggregators;
- Citizen science;
- Data archaeology:
- datasets recovered from scientists’ personal files;
- excel spreadsheets;
- paper documents;
- other formats that would otherwise be lost or inaccessible.
Data product development
EMODnet Biology’s gridded map layers of species abundance for different time windows using geospatial modelling are made available to all users. In addition, we also create spatially distributed data products specifically relevant for Marine Strategy Framework Directive Descriptor 2 (non-indigenous species).
EMODnet Biology is currently working, on the development of the following data products:
- Implementation of a methodology to produce statistically optimized gridded map layers based on Data-Interpolating Variational Analysis (DIVA, ULg);
- Estimation of the accuracy of the gridding procedure by comparison with validated data;
- Complementation of the gridded map of averages with indications of the precision of the result based on the distribution of the basic data used to calculate the products;
- Production of spatial maps of quality indicators relevant for Marine Strategy Framework Directive.
In order to assist our partners, we have developed guidelines on how to create NetCDF files for Biodiversity products. This guidance covers two languages, R and Python, summarises the constraints in using ERDDAP for these type of products and includes a section on CF (Climate and Forecasting) guidance. Due to interest outside the consortium, we have decided to publish the guidance on GitHub so that any interested parties can replicate the procedures with their own products.
Data infrastructure
EMODnet Biology’s data infrastructure and data flow is that of EurOBIS, submitted data undergo a series of quality control procedures before being made available online:
- Metadata;
- Required data fields, including Timestamp, Position, Taxonomy, Provenance, Parameters, Units, among others.
The data infrastructure of EMODnet Biology is able to handle different data protocols and data standards for exchange of marine biodiversity data (e.g. specific data format enabling National Oceanographic Data Centres (NODC’s) to make biological data accessible using the SeaDataNet infrastructure) and adheres to:
- Darwin Core standard used by the Global Biodiversity Information Facility (GBIF) and the Ocean Biodiversity Information System (OBIS);
- OGC Webservices making accessible geospatial data:
- Catalogue Service of the Web (CSW) for metadata resources;
- Web Feature Service (WFS) to allow requests for geographic features across the Web;
- Web Coverage Service (WCS) to allow requests for gridded data across the web;
- Web Map Service (WMS) to allow requests for maps across the web.
- In-house developed web services where the available data are looked at in great detail and a mapping between the data and the Darwin Core Scheme is made allowing to capture as much data and information as possible.
While using controlled vocabularies like:
- World Register of Marine Species (WoRMS ) as taxonomic backbone;
- Marine Regions gazetteer to standardise the geographical locations;
- BODC's NERC Vocabulary Server for the unambiguous identification of measurements or facts associated with the biological occurrences.
Submitted data undergo a series of quality control procedures before being made available online. These procedures focus on:
- Metadata: the data management team checks whether the data and the supplied metadata match and that all necessary fields are filled in correctly and as completely as possible. If important information is missing, a request will be sent to the data provider asking for its completion.
- Required data fields: if the required Darwin Core data fields are not properly filled, a notification will be sent to the data provider to complete them. The data will not be published until all required fields are complete.
- Taxonomy: all taxon names are linked to the World Register of Marine Species (WoRMS). Unmatched taxa are sent back to the data provider for a secondary check-up. Taxa with uncertain identifications are matched to the first suitable higher taxonomic level. Taxon names provided by data originators are stored in the database, as this allows the possibility to go back and revisit the information. When no taxon match can be made, the name is added to an 'annotation list', which keeps track of the editors comments on why a taxon cannot be added to the World Register of Marine Species (see also the section on standards and quality control).
- Geography: all supplied coordinates are converted to the WSG84 coordinate system and expressed as decimal degrees. Furthermore, these coordinates are checked for positioning errors which can include sampling locations on land or in different regions than those included in the metadata information supplied. These errors can be due to accidental swapping of latitude and longitude or related to the use of the minus-sign. All instances found are communicated to the data provider, in order for the necessary corrections to be made.
- Depth: two checks are performed: (1) Is the documented depth-value possible, when compared with the General Bathymetric Chart of the Oceans (GEBCO) and (2) is the documented depth-value possible, when compared with the known depth range of the species?
- Units: if abundance, biomass data or other associated measurements or facts are supplied, the presence of the relevant units is checked. The absence of units, prevents data comparison between different datasets.
Data format
With the EMODnet centralisation several changes impacted the way EMODnet Biology exposes its data. One of the most relevant ones is the data access via the EMODnet Viewer. In Phase V we will continue to work on improving the data access and implementing various filters (e.g. Data source, Time period, Species Traits, Taxonomic grouping as per tender requirements, Parameter filters, etc.) allowing users with a better search capability of the data relevant for their work. The data will be made available as csv files where both the biological occurrences as well as associated parameters are included.
A description of the different fields contained in the output file is described below:
- For all fields not included below, a description can be found in the Darwin Core Reference Guide ;
- Fields used exclusively by EMODnet Biology.
The box 'Biology - Data format' below, contains a list of all terms that are delivered for each option.
| Term | Definition | TDWG URI |
|---|---|---|
| FID | Internal database reference. | |
| id | Unique internal database number. | |
| modified | The most recent date-time on which the record was changed. | purl.org/dc/terms/modified |
| datasetid | An identifier that refers to the metadata record of the dataset in the EMODnet Biology Catalogue. | rs.tdwg.org/dwc/terms/datasetID |
| institutioncode | The name (or acronym) in use by the institution having custody of the object(s) or information referred to in the record. | rs.tdwg.org/dwc/terms/institutioncode |
| collectioncode | The name, acronym, code, or initials identifying the collection or dataset from which the record was derived. | rs.tdwg.org/dwc/terms/collectionCode |
| eventid | An identifier for the set of information associated with an Event (something that occurs at a place and time). May be a global unique identifier or an identifier specific to the dataset. | rs.tdwg.org/dwc/terms/eventID |
| datecollected | A date in full ISO format (YYYY-MM-DDTHH:MM:SS) generated by EMODnet Biology based on the data in the fields yearcollected, monthcollected, daycollected and timeofday. Unknown information is stored as the minimum value (e.g. YYYY-01-01T00:00:00 will be used if only the year is known). | https://dwc.tdwg.org/terms/#dwc:eventDate |
| seasoncollected | The season (winter, spring, summer, autumn) generated by EMODnet Biology based on the data in the fields monthcollected and daycollected. | |
| yearcollected | The year during which the sample or observation occurred. | |
| startyearcollected | For samples or observations that were taken over a duration of time this term contains the start year of the collecting event. If the collection date is uncertain, this term can be used to store the minimum year for of the interval for which there is certainty. | |
| endyearcollected | For samples or observations that were taken over a duration of time this term contains the end year of the collecting event. If the exact year is uncertain, this term can be used to store the maximum year for of the interval for which there is certainty. | |
| monthcollected | The month of the year during which the sample or observation occurred. | |
| startmonthcollected | For samples or observations that were taken over a duration of time this term gives the start month of the collecting event. | |
| endmonthcollected | For samples or observations that were taken over a duration of time this term gives the end month of the collecting event. | |
| daycollected | The day of the year during which the sample or observation occurred. | |
| startdaycollected | For samples or observations that were taken over a duration of time this term gives the start day of the collecting event. | |
| enddaycollected | For samples or observations that were taken over a duration of time this term gives the end day of the collecting event. | |
| timeofday | The time of the day a specimen was collected expressed as decimal hours from midnight (e.g. 12.0 = mid day, 13.5 = 1:30pm). | |
| starttimeofday | For samples or observations that were taken over a duration of time this gives the start time of the day of the collecting event expressed as decimal hours from midnight (e.g. 12.0 = mid day, 13.5 = 1:30pm). | |
| endtimeofday | For samples/observations/record events that were taken over a duration of time this gives the end time of the day of the collecting event expressed as decimal hours from midnight (e.g. 12.0 = mid day, 13.5 = 1:30pm). | |
| timezone | Indicates the time zone for the timeofday measurement. An empty value indicates local time. | |
| waterbody | The name of the water body in which the sample or observation occurred. | rs.tdwg.org/dwc/terms/waterBody |
| country | The name of the country or major administrative unit in which the sample or observation occurred. | rs.tdwg.org/dwc/terms/country |
| stateprovince | The name of the next smaller administrative region than country (state, province, canton, department, region, etc.) in which the sample or observation occurred. | http://rs.tdwg.org/dwc/terms/stateProvince |
| county | The full, unabbreviated name of the next smaller administrative region than stateProvince (county, shire, department, etc.) in which the sample or observation occurred. | rs.tdwg.org/dwc/terms/county |
| recordnumber | An identifier given to the Occurrence at the time it was recorded. Often serves as a link between field notes and an Occurrence record, such as a specimen collector's number. | rs.tdwg.org/dwc/terms/recordNumber |
| fieldnumber | An identifier given to the event in the field. Often serves as a link between field notes and the Event. | rs.tdwg.org/dwc/terms/fieldNumber |
| decimallongitude | The geographic longitude (in decimal degrees, using WGS84 / EPSG:4326) of the geographic center of a Location. Positive values are east of the Greenwich Meridian, negative values are west of it. | rs.tdwg.org/dwc/terms/decimalLongitude |
| startdecimallongitude | For samples or observations that are better represented as line features rather than point features (e.g. extended trawls or transects) this term indicates the starting longitude location from which the specimen was collected. | |
| enddecimallongitude | For samples or observations that are better represented as line features rather than point features (e.g. extended trawls or transects) this term indicates the end longitude location from which the specimen was collected. | |
| decimallatitude | The geographic latitude (in decimal degrees, using WGS84 / EPSG:4326) of the geographic center of a Location. Positive values are north of the Equator, negative values are south of it. | rs.tdwg.org/dwc/terms/decimalLatitude |
| startdecimallatitude | For samples or observations that are better represented as line features rather than point features (e.g. extended trawls or transects) this term indicates the starting latitude location from which the specimen was collected. | |
| enddecimallatitude | For samples or observations that are better represented as line features rather than point features (e.g. extended trawls or transects) this term indicates the end latitude location from which the specimen was collected. | |
| coordinateuncertaintyinmeters | The horizontal distance (in meters) from the given decimallatitude and decimallongitude describing the smallest circle containing the whole of the Location. The value is left empty if the uncertainty is unknown, cannot be estimated, or is not applicable (because there are no coordinates). Zero is not a valid value for this term. | rs.tdwg.org/dwc/terms/coordinateUncertaintyInMeters |
| georeferenceprotocol | A description or reference to the methods used to determine the spatial footprint, coordinates, and uncertainties. | rs.tdwg.org/dwc/terms/georeferenceProtocol |
| minimumdepthinmeters | The minimum distance in metres below the surface of the water at which the collection/record was made; all material collected was at least this deep. Positive below the surface, negative above (e.g. collecting above sea level in tidal areas). | rs.tdwg.org/dwc/terms/minimumDepthInMeters |
| maximumdepthinmeters | The maximum distance in metres below the surface of the water at which the collection/record was made; all material collected was at most this deep. Positive below the surface, negative above (e.g. collecting above sea level in tidal areas). | rs.tdwg.org/dwc/terms/maximumDepthInMeters |
| Occurrenceid | An identifier for the Occurrence. In the absence of a persistent global unique identifier, one is constructed from a combination of identifiers in the record that will most closely make the occurrenceID globally unique. | rs.tdwg.org/dwc/terms/occurrenceID |
| scientificname | The full name of the lowest taxon level that the specimen(s) can be identified as a member of; includes genus, specific epithet, and subspecific epithet (zool.) or infraspecific rank abbreviation, and infraspecific epithet. | rs.tdwg.org/dwc/terms/scientificName |
| scientificnameid | The LSID (life science identifier) which references the scientificname in the World Register of Marine Species. | rs.tdwg.org/dwc/terms/scientificNameID |
| Aphiaid | A link generated by EMODnet Biology based on the scientificnameid that references the scientificname in the World Register of Marine Species. | |
| taxonrank | The taxonomic rank of the scientificName, generated by EMODnet Biology by referencing the scientificnameid in the World Register of Marine Species. | rs.tdwg.org/dwc/terms/taxonRank |
| rankname | The rank name of the most specific name in the scientificName. | |
| scientificnameaccepted | The scientific name of the currently valid or accepted taxon, generated by EMODnet Biology by referencing the scientificnameid in the World Register of Marine Species. | |
| scientificnameauthorship | The authorship information for the scientificName of the currently valid or accepted taxon, formatted according to the conventions of the applicable nomenclaturalCode. | rs.tdwg.org/dwc/terms/scientificNameAuthorship |
| aphiaidaccepted | A link generated by EMODnet Biology based on the scientificnameid that references the currently valid or accepted name for the taxon in the World Register of Marine Species (http://www.marinespecies.org/). | |
| kingdom | The kingdom in which the scientificName of the currently valid or accepted taxon is classified, generated by EMODnet Biology by referencing the scientificnameid in the World Register of Marine Species. | rs.tdwg.org/dwc/terms/kingdom |
| phylum | The phylum in which the scientificName of the currently valid or accepted taxon is classified, generated by EMODnet Biology by referencing the scientificnameid in the World Register of Marine Species. | rs.tdwg.org/dwc/terms/phylum |
| class | The class in which the scientificName of the currently valid or accepted taxon is classified, generated by EMODnet Biology by referencing the scientificnameid in the World Register of Marine Species. | rs.tdwg.org/dwc/terms/class |
| order | The order in which the scientificName of the currently valid or accepted taxon is classified, generated by EMODnet Biology by referencing the scientificnameid in the World Register of Marine Species. | rs.tdwg.org/dwc/terms/order |
| family | The family in which the scientificName of the currently valid or accepted taxon is classified, generated by EMODnet Biology by referencing the scientificnameid in the World Register of Marine Species. | rs.tdwg.org/dwc/terms/family |
| genus | The genus in which the scientificName of the currently valid or accepted taxon is classified, generated by EMODnet Biology by referencing the scientificnameid in the World Register of Marine Species. | rs.tdwg.org/dwc/terms/genus |
| subgenus | The subgenus in which the scientificName of the currently valid or accepted taxon is classified, generated by EMODnet Biology by referencing the scientificnameid in the World Register of Marine Species. | rs.tdwg.org/dwc/terms/subgenus |
| specificepithet | The first or species epithet (species name) of the scientificName of the currently valid or accepted taxon, generated by EMODnet Biology by referencing the scientificnameid in the World Register of Marine Species. | rs.tdwg.org/dwc/terms/specificEpithet |
| infraspecificepithet | The lowest or terminal infraspecific epithet (subspecies name) of the scientificName of the currently valid or accepted taxon, generated by EMODnet Biology by referencing the scientificnameid in the World Register of Marine Species. | rs.tdwg.org/dwc/terms/infraspecificEpithet |
| occurrenceremarks | Comments or notes about the Occurrence. | rs.tdwg.org/dwc/terms/occurrenceRemarks |
| basisofrecord | The specific nature of the data record. | rs.tdwg.org/dwc/terms/basisOfRecord |
| typestatus | A list (concatenated and separated) of nomenclatural types (type status, typified scientific name, publication) applied to the subject. | rs.tdwg.org/dwc/iri/typeStatus |
| catalognumber | An identifier (preferably unique) for the record within the dataset or collection. | rs.tdwg.org/dwc/terms/catalogNumber |
| references | Gives the web address of the page where more information on this particular record (not on the whole dataset) can be found. | purl.org/dc/terms/references |
| recordedby | A list (concatenated and separated) of names of people, groups, or organizations responsible for recording the original Occurrence. The primary collector or observer, especially one who applies a personal identifier (recordNumber), should be listed first. | rs.tdwg.org/dwc/terms/recordedBy |
| identifiedby | A list (concatenated and separated) of names of people, groups, or organizations who assigned the Taxon to the subject. | rs.tdwg.org/dwc/terms/identifiedBy |
| yearidentified | The year on which the subject was identified as representing the Taxon. | |
| monthidentified | The month on which the subject was identified as representing the Taxon. | |
| dayidentified | The day on which the subject was identified as representing the Taxon. | |
| preparations | A preparation or preservation method for a specimen. | http://rs.tdwg.org/dwc/iri/preparations |
| samplingeffort | The amount of effort expended during an Event. | rs.tdwg.org/dwc/terms/samplingEffort |
| samplingprotocol | The method or protocol used during an Event. | rs.tdwg.org/dwc/iri/samplingProtocol |
| the_geom | A Well-Known Text (WKT) representation of the shape (footprint, geometry) that defines the dcterms:Location. A dcterms:Location may have both a point-radius representation (see dwc:decimalLatitude) and a footprint representation, and they may differ from each other. | https://dwc.tdwg.org/terms/#dwc:footprintWKT |
| eurobis_unique_positions_id | ||
| julianday | ||
| startjulianday | ||
| endjulianday | ||
| coordinateprecision_category | ||
| start_endcoordinateprecision | ||
| boundingbox | ||
| minimumelevation | The lower limit of the range of elevation (altitude, usually above sea level), in meters. | https://dwc.tdwg.org/terms/#dwc:minimumElevationInMeters |
| maximumelevation | The upper limit of the range of elevation (altitude, usually above sea level), in meters. | https://dwc.tdwg.org/list/#dwc_maximumElevationInMeters |
| depthrange | The original description of the depth below the local surface. | https://dwc.tdwg.org/list/#dwc_verbatimDepth |
| temperature | A list of additional measurements, facts, characteristics, or assertions about the record. Meant to provide a mechanism for structured content. | http://rs.tdwg.org/dwc/terms/#dynamicProperties |
| individualcount | The number of individuals present at the time of the dwc:Occurrence. | https://dwc.tdwg.org/list/#dwc_individualCount |
| observedweight | A list of additional measurements, facts, characteristics, or assertions about the record. Meant to provide a mechanism for structured content. | http://rs.tdwg.org/dwc/terms/#dynamicProperties |
| previouscatalognumber | A list (concatenated and separated) of previous or alternate fully qualified catalog numbers or other human-used identifiers for the same dwc:Occurrence, whether in the current or any other data set or collection. | https://dwc.tdwg.org/list/#dwc_otherCatalogNumbers |
| relationshiptype | The relationship of the subject (identified by dwc:resourceID) to the object (identified by dwc:relatedResourceID). | https://dwc.tdwg.org/list/#dwc_relationshipOfResource |
| relatedcatalogitem | A list (concatenated and separated) of identifiers of other dwc:Occurrence records and their associations to this dwc:Occurrence. | https://dwc.tdwg.org/list/#dwc_associatedOccurrences |
| samplesize | A list of additional measurements, facts, characteristics, or assertions about the record. Meant to provide a mechanism for structured content. | http://rs.tdwg.org/dwc/terms/#dynamicProperties |
| qc | ||
| datageneralizations | Actions taken to make the shared data less specific or complete than in its original form. Suggests that alternative data of higher quality may be available on request. | https://dwc.tdwg.org/list/#dwciri_dataGeneralizations |
| eventremarks | Comments or notes about the Event. | http://rs.tdwg.org/dwc/terms/eventRemarks |
| locationremarks | Comments or notes about the Location. | http://rs.tdwg.org/dwc/terms/locationRemarks |
| associatedsequences | A list (concatenated and separated) of identifiers (publication, global unique identifier, URI) of genetic sequence information associated with the Occurrence. | http://rs.tdwg.org/dwc/terms/associatedSequences |
| associatedmedia | A list (concatenated and separated) of identifiers (publication, global unique identifier, URI) of media associated with the Occurrence. | http://rs.tdwg.org/dwc/terms/associatedMedia |
| organismid | An identifier for the Organism instance (as opposed to a particular digital record of the Organism). May be a globally unique identifier or an identifier specific to the dataset. | http://rs.tdwg.org/dwc/terms/organismID |
| geodeticdatum | The ellipsoid, geodetic datum, or spatial reference system (SRS) upon which the geographic coordinates given in decimalLatitude and decimalLongitude as based. | http://rs.tdwg.org/dwc/terms/geodeticDatum |
| dynamicproperties | A list of additional measurements, facts, characteristics, or assertions about the record. Meant to provide a mechanism for structured content. | http://rs.tdwg.org/dwc/terms/dynamicProperties |
| identificationverificationstatus | A categorical indicator of the extent to which the taxonomic identification has been verified to be correct. | http://rs.tdwg.org/dwc/terms/identificationVerificationStatus |
| identificationreferences | A list (concatenated and separated) of references (publication, global unique identifier, URI) used in the Identification. | http://rs.tdwg.org/dwc/terms/identificationReferences |
| organismquantity | A number or enumeration value for the quantity of dwc:Organisms. | https://dwc.tdwg.org/list/#dwc_organismQuantity |
| organismquantitytype | The type of quantification system used for the quantity of organisms. | http://rs.tdwg.org/dwc/iri/#organismQuantityType |
| samplesizevalue | A numeric value for a measurement of the size (time duration, length, area, or volume) of a sample in a sampling event. | http://rs.tdwg.org/dwc/terms/sampleSizeValue |
| samplesizeunit | The unit of measurement of the size (time duration, length, area, or volume) of a sample in a sampling event. | http://rs.tdwg.org/dwc/iri/sampleSizeUnit |
| materialsampleid | A physical result of a sampling (or subsampling) event. In biological collections, the material sample is typically collected, and either preserved or destructively processed. | http://rs.tdwg.org/dwc/terms/MaterialSample |
| taxonconceptid | An identifier for the taxonomic concept to which the record refers - not for the nomenclatural details of a taxon. | http://rs.tdwg.org/dwc/terms/taxonConceptID |
| seasonid | ||
| dataprovider_id | Internal mapping to the data provider. | |
| parameter | ||
| parameter_value | ||
| parameter_group_id | ||
| parameter_measurementtypeid | ||
| parameter_bodcterm | ||
| parameter_bodcterm_definition | ||
| parameter_standardunit | ||
| parameter_standardunitid | ||
| parameter_imisdasid | ||
| parameter_ipturl | ||
| parameter_original_measurement_type | ||
| parameter_original_measurement_unit | ||
| parameter_conversion_factor_to_standard_unit | ||
| event | An action that occurs at some location during some time. | http://rs.tdwg.org/dwc/terms/Event |
| event_type | The nature of the dwc:Event. | https://dwc.tdwg.org/list/#dwc_eventType |
| event_type_id | ||
| occurrenceid_unique | Internal generated number. | |
| parameter_standardunitid | ||
| parameter_imisdasid |
Communication
In this section you will find thematic-specific communication material, and a direct link to the video gallery.
Leaflets